MOLECULAR BASIS OF INHERITANCE:
CHAPTER AT A GLANCE
THE DNA
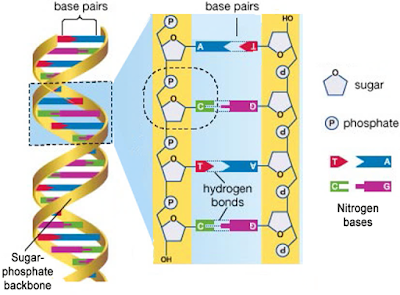
DNA & RNA are polynucleotides (polymer of nucleotides).
Nucleoside= A nitrogen base + pentose sugar (by N-glycosidic bond).
Nucleotide= A nitrogen base + A pentose sugar (ribose in RNA & deoxyribose in DNA) + a phosphate group.
Nitrogen bases are 2 types:
- Purines: Adenine (A) and Guanine (G).
- Pyrimidines: Cytosine (C), Thymine (T) & Uracil (U).
Phosphodiester bond= Bond b/w sugar & phosphate.
Erwin Chargaff’s rule:
In DNA, the proportion of A is equal to T and the proportion of G is equal to C.
[A] + [G] = [T] + [C] or [A] + [G] / [T] + [C] =1
DNA (-ve charge) is wrapped around histone octamer (+ve charge) to give nucleosome.
[A] + [G] = [T] + [C] or [A] + [G] / [T] + [C] =1
PACKAGING OF DNA HELIX
DNA (-ve charge) is wrapped around histone octamer (+ve charge) to give nucleosome.
Higher level packaging of chromatin needs non-histone chromosomal (NHC) proteins.
Chromatin has 2 forms:
- Euchromatin: Loosely packed, light stained and transcriptionally active region.
- Heterochromatin: Densely packed, dark stained and inactive region.
THE SEARCH FOR GENETIC MATERIAL
Griffith’s Transforming Principle experiment:
- S-strain → Inject into mice → Mice die
- R-strain → Inject into mice → Mice live
- S-strain (Heat killed) → Inject into mice → Mice live
- S-strain (Hk) + R-strain (live) → Inject into mice → Mice die
Biochemical characterization of transforming principle:
By Avery, MacLeod & McCarty.
They purified biochemicals from heat killed S cells using suitable enzymes.
Digestion of DNA with DNase inhibited transformation. It proves that DNA was the transforming principle.
Hershey-Chase Experiment (Blender Experiment):
Bacteriophage viruses + radioactive phosphorus (P32) → radioactive DNA → Infected with E. coli.
Bacteriophage viruses + radioactive sulphur (S35) → radioactive protein → Infected with E. coli.
Blending to remove virus particles from bacteria.
Centrifugation to separate lighter virus particles from heavier bacterial cells.
Bacteria infected with viruses having radioactive DNA were radioactive. i.e., DNA had passed from the virus to bacteria.
Bacteria infected with viruses having radioactive proteins were not radioactive. i.e., proteins did not enter the bacteria from the viruses. This proves that DNA is the genetic material.
PROPERTIES OF GENETIC MATERIAL
(DNA v/s RNA)
- Replication.
- Chemical and structural stability.
- Show mutations for evolution.
- Express as Mendelian Characters.
- Double stranded
- Presence of thymine
- Absence of 2’-OH in sugar
- Single stranded
- Presence of Uracil
- Presence of 2’-OH in sugar
CENTRAL DOGMA OF MOLECULAR BIOLOGY
It is proposed by Francis Crick.
DNA REPLICATION
Replication is the copying of DNA from parental DNA.
Watson & Crick proposed Semi-conservative model of replication.
Messelson & Stahl’s Experiment:
- They grew E. coli in 15NH4Cl medium (15N = heavy isotope). As a result, new heavy DNA (15N DNA) formed.
- Heavy DNA can be distinguished from normal DNA (light DNA or 14N DNA) by centrifugation in cesium chloride density gradient.
- E. coli cells from 15N medium were transferred to 14N medium. In next generation, density of DNA was intermediate b/w 15N DNA & 14N DNA. i.e., one strand is old (15N) and one strand is new (14N).
DNA replication starts at a point called origin.
DNA replicates in the 5’→3’ direction.
Deoxyribonucleoside triphosphates act as substrate.
2 strands unwind and separate to form replication fork.
In presence of DNA polymerase, nucleotides join to form new strand.
One strand undergoes Continuous synthesis.
Other strand undergoes discontinuous synthesis forming Okazaki fragments. They join to form a new strand by DNA ligase.
TRANSCRIPTION
Formation of RNA from one strand of the DNA.
3’-ATGCATGCATGCATGCATGCATGC-5’ template strand.
5’-TACGTACGTACGTACGTACGTACG-3’ coding strand.
In transcription, both strands are not copied because
- The code for proteins is different in both strands.
- 2 RNA molecules form double stranded RNA.
◦ A promoter: Binding site for RNA polymerase.
◦ Structural gene: Region b/w promoter and terminator.
◦ A terminator: The site where transcription stops.
Structural gene in a transcription unit is 2 types:
- Monocistronic structural genes (split genes): Seen in eukaryotes. It contains exons and introns.
- Polycistronic structural genes: Seen in prokaryotes. Here, there are no split genes.
- Initiation: RNA polymerase binds at promoter site → unwinding of DNA. An initiation factor (σ factor) in RNA polymerase initiates RNA synthesis.
- Elongation: RNA chain is synthesized in 5’-3’ direction. Activated ribonucleoside triphosphates are added.
- Termination: A termination factor (ρ factor) binds to the RNA polymerase and terminates the transcription.
1. There are 3 RNA polymerases: RNA polymerase I, II & III.
2. Primary transcripts (hnRNA) contain exons & introns. To remove introns, it undergoes the following processes and become mRNA:
- Splicing: Introns are removed and exons are joined.
- Capping: Methyl guanosine triphosphate (cap) is added to the 5’ end of hnRNA.
- Tailing (Polyadenylation): Adenylate residues (200-300) are added at 3’-end.

GENETIC CODE
It is the sequence of nucleotides (nitrogen bases) in mRNA that contains information for protein synthesis.
Salient features of genetic code:
- Triplet code.
- 61 codons code for amino acids. UAA, UAG & UGA are stop codons (Termination codons).
- Genetic code is universal.
- No punctuations b/w adjacent codons.
- An amino acid is coded by many codons. So the code is degenerate.
- AUG has dual functions: codes for Methionine + initiator codon.
TYPES OF RNA
- mRNA (messenger RNA): Provide template for translation (protein synthesis).
- rRNA (ribosomal RNA): catalytic role during translation.
- tRNA (transfer RNA): Adapter molecule. Brings amino acids for protein synthesis and reads the genetic code. It has an Anticodon loop & an amino acid acceptor end.
TRANSLATION (PROTEIN SYNTHESIS)
- Charging (aminoacylation) of tRNA: Amino acids are activated (amino acid + ATP) + tRNA.
- Initiation: Ribosome binds to mRNA at the start codon (AUG). So the initiator tRNA (with methionine) binds. Its anticodon (UAC) recognises start codon AUG.
- Elongation: Second aminoacyl tRNA binds to ribosome. Its anticodon binds to second codon. A peptide bond is formed between first and second amino acids. This process continues.
- Termination: It occurs when a release factor binds to stop codon.
REGULATION OF GENE EXPRESSION
Levels of gene expression in eukaryotes:
- Transcriptional level
- Processing level
- Transport of mRNA from nucleus to the cytoplasm
- Translational level.
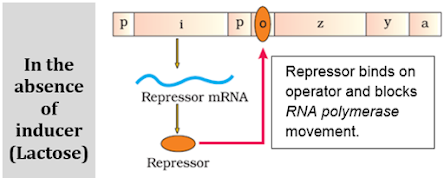
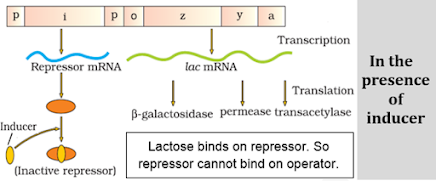
a) A regulatory or inhibitor (i) gene: Codes for repressor protein.
b) 3 structural genes:
- z gene: Codes for b galactosidase. It hydrolyses lactose to galactose and glucose.
- y gene: Codes for permease. It increases permeability of the cell to lactose.
- a gene: Codes for a transacetylase.
HUMAN GENOME PROJECT (HGP)
First mega project for sequencing of nucleotides and mapping of all genes in human genome.
Goals of HGP:
- Identify all the genes in DNA.
- Sequencing of 3 billion base pairs of human DNA.
- Store this information in databases.
- Improve tools for data analysis.
- Transfer related technologies to other sectors.
- Address the ELSI.
- Expressed Sequence Tags (ESTs): Focused on identifying all the genes that are expressed as RNA.
- Sequence annotation: Sequencing whole genome.
Isolate DNA from a cell → Convert into random fragments → Clone in a host using vectors → Sequencing of fragments using Automated DNA sequencers (Frederick Sanger method) → Arrange the sequences based on overlapping regions→ Alignment of sequences using computer programs.
Salient features of Human Genome:
- Contains 3164.7 million bases & 30,000 genes.
- 99.9% nucleotide bases are same in all people.
- Chromosome I has most genes (2968) and Y has the fewest (231).
- Major portion of genome is made of Repeated (repetitive) sequences.
- 1.4 million locations have single-base DNA differences. They are called SNPs (Single nucleotide polymorphism or ‘snips’).
DNA FINGERPRINTING (DNA PROFILING)
Technique to identify similarities & differences of the DNA fragments of 2 individuals.
It is developed by Alec Jeffreys.
Basis of DNA fingerprinting:
DNA carries non-coding repeated sequences called variable number tandem repeats (VNTR).
VNTR is specific in each person.
Steps (Southern Blotting Technique):
- Isolation of DNA.
- Digestion of DNA by restriction endonucleases.
- Separation of DNA fragments by gel electrophoresis.
- Transferring (blotting) DNA fragments to nitrocellulose or nylon membrane.
- Hybridization by radioactive VNTR probe.
- Detection of hybridized DNA by autoradiography.
- Forensic tool to solve paternity, rape, murder etc.
- For the diagnosis of genetic diseases.
- To determine phylogenetic status of animals.






superb
ReplyDeleteOne of the best study material
ReplyDelete